Global Journal of Zoology
Chromatin Remodeling Complexes: The Regulators of Genome Function
Shashibhal M Pandey*
Cite this as
Pandey SM (2016) Chromatin Remodeling Complexes: The Regulators of Genome Function. Glob J Zool 1(1): 007-013. DOI: 10.17352/gjz.000003Genome in eukaryotes is large enough to be accommodated in tiny nucleus. It is required to achieve high degree of compaction for getting into the nucleus. Compaction is achieved by folding the DNA in the form of chromatin. But chromatin acts as general repressor for the entire genomic functions. Therefore, it requires being selectively unpacked for gene expression. However this packing and unpacking of chromatin need to temporally and spatially regulated for differential regulation of genomic functions like DNA replication, repair, recombination and transcription. Chromatin remodeling factors regulate structure and function of chromatin in time and space to facilitate various genomic functions. Chromatin remodeling complexes can be broadly categorized into those that carry out remodeling by utilizing energy from ATP hydrolysis and those that covalently modify chromatin proteins and thus bring about permanent yet reversible alteration in the chromatin structure.
Introduction
Living organisms are improbable steady-state systems. They exist in environment that changes from seconds to minute, day to day and over the years and centuries. If we ask what enables them to continue on this unlikely course, the answer will be “information” that they inherit in their genetic material, which allows them to take actions to prevent death. They can do this on various scales. Every organism is an agent selecting from time to time what is best to do in the changing circumstances. An organism can choose ‘wisely’ because its ‘genome’ provides it with receptors tuned to respond to changes that are likely to occur. Such a diploid genome in case of human is about 2 meters long and must be compacted to fit into a nucleus with diameter that is about 200,000 fold smaller. And the situation is quite similar in other eukaryotes. Eukaryotic cells have solved this packaging problem by folding their DNA along with protein into a highly compacted structure called ‘chromatin’ [1].
Chromatin: A solution and the problem
All the chromatin proteins may be divided into two categories; histones and non-histone protein. This distinction is based on unique characteristics and functions of histones. Their relative amounts and stoichiometry with respect to DNA are nearly constant throughout the eukaryotic kingdom. Histone proteins form a core around which DNA is tightly wrapped forming ‘nucleosome’, the structural unit of chromatin. Biochemical and genetic experiments over the past two and a half decade have confirmed that the organization of eukaryotic DNA in chromatin exerts a general repressive effect on replicative and transcriptional processes [2]. Therefore after solving the problem of packaging the genome, chromatin structure give rise to another problem of accessibility of the genome by various processes requiring DNA as the substrate.
Replicative and transcriptional processes therefore require the chromatin to be differentially unpacked and subsequently packaged, with minute temporal and spatial precision. One of the most intriguing phenomena related to chromatin structural variability is the presence of two morphologically different types of chromatin within a single inter-phase nucleus: the dispersed euchromatin and condensed heterochromatin. The nature of replicative and transcriptional mechanics in vivo poses a tough challenge to our understanding, about how the chromatin condensation (and decondensation) is accomplished. Even though we know very little about the mechanism and nature of higher-order chromatin folding and unfolding, the chromatin changes at a simpler (nucleosomal) level of organization have been unraveled [1]. The X- ray crystal structure of both the histone octamer and the nucleosome core particles have been obtained at high resolution. During the past several years these structures have served as the primary basis for interpreting nucleosome function in chromatin fibers. The nucleosome [3-5] is made up of an octamer of four core-histones H2A, H2B, H3 and H4 around which about 147 bp of DNA wraps 1.65 left handed superhelical turns, assuming a molecular mass of ~206 kDa.
Different DNA binding proteins are affected differentially by the nucleosomal DNA. A common finding too many DNA binding proteins is that if its cognate DNA binding site is located close to nucleosome border then it is more accessible than the same DNA site located close to the nucleosomal dyad [6]. One extreme is nuclear factor NF1 which has 100-300 fold reduced affinity for its nucleosomal site compared to free DNA, independent of translational and rotational positioning. It should be noted however, that a rotationally unfavorable positioned DNA element where the factor binding surface, forming the base specific protein contacts, is facing towards the histone octamer, in most cases, usually has drastically lower accessibility, than if this surface is positioned away from the histone surface [7].
Nucleosomes are not structurally inert but instead undergo several conformational transitions that are dynamic and likely to be important in vivo. At molecular level nucleosomal DNA exist in dynamic equilibrium between wound and unwound state to the histone octamer [8,9]. This dynamic behavior exposes DNA sites with a probability of 1 in 103 to 105 as one moves from periphery towards the centre, so the apparent DNA binding affinities of many trans acting factors for nucleosomal DNA simply will be reduced by 103 to 105 fold compared with the affinities of these factors for the same site on naked DNA. Thus, although the time averaged fraction of nucleosomes with exposed binding site is small, factors having sufficiently high affinity for naked DNA and/or present in locally high concentration will still be able to bind to their cognate element in chromatin, as dictated by laws of mass action.
Chromatin remodeling complexes: A solution to the problem
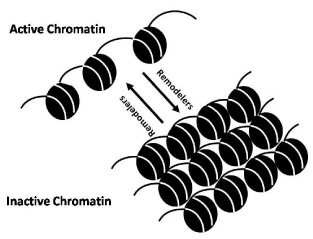
It is evident that structure of nucleosome described above renders nucleosomal DNA less accessible. One molecular solution to the problem of chromatin restructuring is provided by the activities of chromatin remodeling factors [Figure 1]. Two classes of chromatin remodeling factors have been described. First class of chromatin remodeling factors in includes protein complexes that bring about alteration in the chromatin structure by covalently modifying histones. Whereas second class remodeling complexes are of molecular motors, the ATP-dependent chromatin remodeling factors [10].
ATP-independent chromatin remodeling
ATP-independent chromatin remodeling is brought about by factors that are responsible for posttranslational, covalent modifications in various histones. Large number of elegant review articles published recently, provide in depth analysis of composition and its functional implication of histone modifying complexes [11]. However, in order to stay focused on the theme of this thesis, chromatin remodeling by various modifications of chromatin, have been briefly addressed below.
Chromatin remodeling by chromatin modifications
The core histones that make up nucleosome are subject to more than 100 different post translational modifications: acetylation [11], methylation [12] phosphorylation, ADP ribosylation and ubiquitilation [11,13]. These occur primarily at specific positions within the non-globular amino-terminal histone tails which protrude from the core of the nucleosome (explained above). Since they were discovered in 1960’s histone modifications have been predicted to affect all aspects of chromosome biology, including transcription, replication, recombination and condensation, by affecting chromosome structure and by recruiting specific chromatin-binding proteins.
There have seen considerable progress in understanding of acetylation and methylation of lysine residues in the histones. On a genome wide basis, histone H3 K4 tri-methylation and H3K9 acetylation are associated with active transcriptional start sites. Phosphorylated H2A.X foci mark sites of DNA damage and methylation of Histone H3 Lys9 recruits chromodomain-containing proteins, such as heterochromatin protein 1 (HP1) [14]. Acetylation of lysine residue in histone octamer almost always correlates with chromatin accessibility and transcriptional activity; and that the functional importance of acetylation depends completely on the accuracy and efficiency of the reverse reaction, histone deacetylation. One of the way in which modifications such as acetylation affect transcription is based on the recruitment of activators due to recognition via modification binding domain for example acetylation of histones recruits bromodomain-containing proteins [15-17]. However, recent advances in the methylation related studies indicate that lysine methylation can have different effects depending on which residue is modified. Methylation, in particular, was linked to the regulation of gene expression and chromatin conformation [18,19]. Most modifications are dynamic. Although histone methylation was long considered irreversible, the recent identification of numerous site-specific histone demethylases provides compelling evidence that this modification is dynamically regulated [20]. A flurry of recent studies has offered glimpses into the specific biological roles of the histone modifying enzymes and their potential connections to human diseases [20]. Of all the enzymes that modify histones, methyl transferase and kinases are the most specific.
There are two characterized mechanisms for the function of modifications. The first is disruption of contacts between nucleosomes in order to unravel chromatin and second is the recruitment of non-histone proteins. Second one is the most characterized till date. Thus depending upon the composition of modifications (histone code) on a given histone a set of proteins are encouraged to bind or are occluded from chromatin. These proteins carry with them enzymatic activities (i.e. remodeling ATPases) that further modify chromatin. The need to recruit an ordered series of enzymatic activities comes from the fact that processes regulated by modifications (transcription, replication, repair) have several steps. Each one of these steps may require a distinct type of chromatin remodeling activity and a different set of modifications to recruit them [11].
ATP dependent chromatin remodeling
ATP dependent chromatin remodeling is brought about by the factors called remodelers. Remodelers are DNA dependent motors that utilize energy derived from ATP hydrolysis to non-covalently alter this structure [21,22]. These enzymes are member of a diverse group of proteins named (SWI/SNF) after the archetypal S. Cerevisiae Snf2 proteins; the Snf2 family. Multiple members of this protein family are present in the sequenced genomes of eukaryotes, of which the chromatin remodeling enzymes form distinct sub groupings [23]. The crystal structure of catalytic domains of the two Snf2 related proteins highlight structural similarities with the RecA domain found in the range of helicasess [24]. Snf2 proteins use the energy of ATP hydrolysis to alter the histone DNA interaction. However, unlike bona-fide helicases, the action of chromatin remodeling enzymes are not generally associated with separation of DNA strands.
Remodelers can in vitro mediate (a) nucleosome sliding, in which the position of nucleosome on DNA changes, (b) the creation of a remodeled state, in which DNA becomes more accessible but histones remain bound, (c) complete dissociation of histone and DNA, or (d) histone replacement with variant histones (for a detailed discussion see below). At the same time, ATP dependent remodelers work in conjunction with histone chaperones and histone modifying enzymes [25].
Currently, four different classes of ATP-dependent remodeling complexes can be recognized: SWI/SNF, ISWI, Mi-2, and Ino80. Each class is defined by the presence of a distinct ATPase [10].
SWI/SNF group
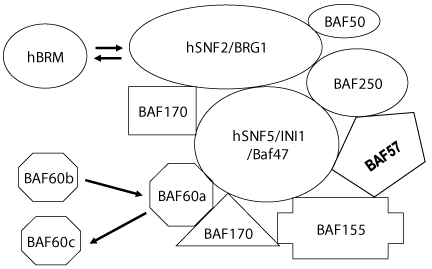
Historically, it was the discovery of yeast SWI/SNF complexes in the mid-1980s initiated spurt in studies of chromatin remodeling. First chromatin remodeling complex was purified from yeast. It is product of five SWI and SNF gene (SWI1, SWI2/SNF2, SWI3, SNF5 and SNF6) were found to be constituents of a 2 MDa complex [26,27] named SWI/SNF complex. Later on affinity-purified complex contained, in addition to SWI1, SWI2/SNF2, SWI3, SNF5 and SNF6, five more then-unknown proteins with molecular weights of 78, 68, 50, 47 & 25 kDa [28].
All prototype SWI/SNF-type complexes studied so far contain a minimal structural and functional core composed of four evolutionarily-conserved subunits: homolog of yeast proteins SWI2/SNF2 (the ATPase, major catalytic subunit), SNF5, SWI3 and SWP73 [29,30]. The complex has an ATPase activity that is stimulated by DNA (~30 fold) or by nucleosomes (~40 fold) [28]. Functional characterizations of the complex revealed that it could stimulate binding of GAL4 (and GAL4 derivatives) to nucleosomal binding sites in presence of ATP. In a mutated complex, wherein the SWI2/SNF2-NTP binding motif is rendered non-functional by a point mutation (K798→A), fails to stimulate activator binding to nucleosomes. This suggests that the ATPase activity of SWI2/SNF2 is essential for the SWI/SNF function, but is not needed for structural assembly of the complex. The complex was found (i) to bind DNA in a sequence-non-specific manner with preference for four-way junction DNA, (ii) to interact with DNA through the minor groove, and (iii) to induce positive supercoiling in relaxed plasmids in the presence of ATP [31]. The complex was however redundant when multiple transcription factors bind to nucleosomes in vitro [32]. Reportedly, the yeast SWI/SNF complex (i) disrupted a nucleosome in the presence of ATP, and (ii), persistently remodeled a specific GAL4-binding site-containing nucleosome along an array of nucleosomes in presence of ATP and GAL4, and (iii) evicted histones from activator-interactive nucleosomes in the presence of an activator [33]. In addition, the complex was found to slide nucleosome along a longer DNA fragment [34]. The available data indicate that the subunits have specific roles in determining the range of targets and biological functions of the complexes.
SWI/SNF group of remodelers can be further subdivided into two distinct highly conserved subclasses. One subfamily comprises yeast SWI/SNF, Drosophila BAP (Brm associated proteins) and mammalian BAF complex; whereas the second family includes yeast RSC, Drosophila PBAP, and mammalian PBAF complexes [Figure 2]. Chromatin remodeling activity although well-established across the animal phyla has also been reported in plant [35].
In animals, the subunits of SWI/SNF complexes are involved in key developmental pathways at both early and later stages of the life cycle. The mechanisms underlying this involvement include: (i) direct interactions with promoter- or enhancer-binding proteins [36], (ii) mediation of glucocorticoid- and stress-induced apoptosis [37], (iii) contribution to genomic recombination, e.g., during T-cell differentiation [38], (iv) physical interactions with components of signaling pathways, e.g., interactions of BRG1 ATPase with inositol, promoted association of SWI/SNF subunits with chromatin [39,40] and (v) promoting cell-cycle arrest, either by down regulation of E2F target genes, like cyclin E, or up-regulation of cyclin-dependent kinase inhibitors [41].
ISWI group
The first member of this growing group of chromatin remodeling complexes, dNURF, and dCHRAC were identified in Drosophila embryo extract using in vitro assays for activities allowing transcription factor access to sites in nucleosomal arrays [42,43]. Later multiple additional remodelers belonging to this group were identified in yeast [44], humans [45,46], mouse [47] and Xenopus [48]. The ATPase subunit of this group of remodeling complex was named Imitation switch (ISWI) because of its similarity to SWI2 ATPase in the SNF2 subfamily DEAD/H helicases. Characteristic of ISWI type ATPase is the presence of a SWI3, ADA2, N-CoR and TFIIIB (SANT) domain and absence of a bromodomain [49]. These remodeling complexes have an ATPase subunit that belongs to the SWI2/SNF2 subfamily of DEAD/H helicases [50]. SANT-like domains in the catalytic subunit have been proposed to interact with histone tails [51,52]. The complexes in this group are relatively smaller (300-800kDA) and contain fewer subunits ranging from 2-4 as compared with larger complexes in the SNF2, CHD, and INO80 subfamilies which contain upto 15 subunits and are often ~2MDa.
CHD/Mi2 Group
In addition to having a Swi2/Snf2-like helicase/ATPase domain, members of the CHD subfamily also contain a chromo (chromatin organization modifier) domain and a DNA-binding domain [53]. Chromo domains are found in a number of proteins, including many that have the ability to interact with heterochromatin, such as Droshophila Polycomb, Drosophila HP1, S. pombe Clr4 histone methyltransferase (HMTase), S. pombe Swi6, and mammalian SUV39H1 HMTase. The mechanism by which chromo domains are brought to heterochromatin is unclear, but it may involve binding to methylated histones. Recently, chromo domains were found to act as recognition motifs for methylated lysine-9 of histone H3 [14]. Chromo domains have also been shown to interact with RNA as well as to self-associate with one another [54]. Many complexes that contain a CHD family member and that display both histone deacetylase and ATP-dependent nucleosome disruption activities were purified from both humans and Xenopus laevis. In humans, this complex was individually identified by three different laboratories, and named NURD (nucleosome remodeling and histone deacetylation), NuRD, and NRD (nucleosome remodelling and deacetylating) [55-57]. CHD4/Mi-2h and CHD3/Mi-2a are highly related proteins that are autoantigens in the human disease dermatomyositis. These proteins are ATPases and presumably lead to the ATP-dependent chromatin-remodeling activity of NURD complexes, and in fact recombinant human Mi-2 was found to have ATPase activity comparable to intact NuRD complex [58].
A Mi-2 homolog also exists in Drosophila, named dMi- 2 that exists in a large complex, similar to its human and Xenopus counterparts, but this complex is much less characterized. It does seem to contain histone deacetylase activity. Some striking differences between recombinant dMi-2 and ISWI were found. The ATPase activity of dMi-2’s ATPase is only stimulated by nucleosomes. Furthermore, dMi-2 was able to bind nucleosome cores (which presumably display no free DNA), and dMi-2 move histone octamers in opposite directions in a sliding assay, suggesting that in contrast to ISWI remodeling complexes aMi-2 use different mechanisms of nucleosome mobilization [53]
A complex highly homologous to the NURD complexes was isolated from Xenopus egg extracts that demonstrate histone deacetylase activity along with nucleosome-stimulated ATPase activity [59,60].
INO80.com
The INO80 gene (YGL150C) was identified in a genetic screen for mutants affecting inositol biosynthesis [61]. The product of this gene is highly related to the DNA-dependent ATPases in the SNF2/SWI2 superfamily of chromatin remodeling complexes. The Wu group purified and characterized the INO80 complex.
The purified INO80 complex contains 15 principal subunits with roughly equivalent stoichiometry except for Rvb1 and Rvb2, which show 6:1 stoichiometry with other polypeptides [62]. INO80 complex is highly conserved in human INO80 complex contains orthologs of Ino80, Rvb1, Rvb2, Arp4, Arp5, Arp8, Ies2 and Ies6, as well as five unique subunits [63].
The in vitro biochemical studies showed that the INO80 complex has DNA-dependent ATPase activity, as well as 3’–5’ helicase activity [62]. INO80.com was also found to be able to bind to free DNA with an apparent binding constant (~10 nM), which is comparable to that of SWI/SNF [64]. The INO80.com participates in multiple DNA repair pathways by its nucleosome remodeling ability and by regulating the accessibility of DNA repair proteins around the DSB site. Though the INO80 complex has been previously shown to play an important role in transcription, the recent finding on the roles of INO80 complex during the DNA damage response emphasizes the notion that chromatin remodeling complexes can be involved in distinctly different cellular processes [65]. DNA. INO80-C and RSC remodelers play important role in nucleosome eviction at DSB (double stranded breaks) and facilitate Rad51 binding [66]. Additionally INO80 and RSC both remodelers appear to contribute to end resection.
Conclusion and Future Perspective
This review discussed the problems posed by large size of genome in eukaryotes and its solution provided by packaging of the genome in the form of chromatin. However his packaging makes genome inaccessible to different factors that require DNA as substrate. For genome functions like replication, repair, recombination and transcription, it requires differential unpack aging of genomic chromatin. Eukaryotic cells have solved this problem by evolving chromatin remodeling enzymes and processes that regulate chromatin structure and differential gene expression in vivo. Many chromatin remodeling complexes from yeast, Drosophila and human have been purified and characterized. Yet there are left many systems like bird and fishes where a remodeling complex is yet to be purified. On one hand it can help in bridging the gap that exist between identified remodeling complexes, while on the other hand it may lead to identification of novel subunits, their functions and regulation, that will add to the dynamicity of these complexes. Finally in human health chromatin remodeling can be used for diagnosis and prognosis of traits like current and the time course activity of disease.
I gratefully acknowledge support lab resources from Prof. M.M. Chaturvedi, of Delhi University. I am also thankful to Mumbai University for providing financial assistance for minor research project on cloning and expression of Brg1 protein.
- Lewin B (2008) Genes IX. Ed. Jones and Barlwtt publishers, Inc. Link: https://goo.gl/NUrtnv
- Felsenfeld G (1996) Chromatin unfolds. Cell 86: 13-19. Link: https://goo.gl/WyHPi8
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature 389: 251-260. Link: https://goo.gl/Q2d3B6
- Wolffe AP, Kurumizaka H (1998) The nucleosome; A powerful regulator of transcription. Prog Nucleic Acid Res Mol Biol 61: 379-422. Link: https://goo.gl/7mxMx4
- Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 96: 285-294. Link: https://goo.gl/g9W4X9
- Li Q, Wrange O (1993) translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev 7; 2471–2482. Link: https://goo.gl/molniW
- Li Q, Wrange O (1995) Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol Cell Biol 15: 4375–4384. Link: https://goo.gl/3QQM7Y
- Anderson JD, Widom J (2000) Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 296: 979–987. Link: https://goo.gl/UOXJjb
- Polach KJ, Lowary PT, Widom J (2000) Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 298: 211-23. Link: https://goo.gl/sbWHzf
- Imbalzano A, Xiao H (2005) Functional properties of ATP dependent chromatin remodeling enzymes. Advances in Protein Chem 67: 157-179. Link: https://goo.gl/u6x18b
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693-705. Link: https://goo.gl/EdLRfL
- Rice JC, Allis CD (2001) Gene regulation: Code of silence. Nature 414: 258-261. Link: https://goo.gl/JzTAAh
- Wolffe AP, Guschin D (2000) Review: Chromatin structural features and targets that regulate transcription. J Struct Biol 129: 102-122. Link: https://goo.gl/daRGn9
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116– 120. Link: https://goo.gl/Ng7MOA
- Dhalluin C1, Carlson JE, Zeng L, He C, Aggarwal AK, et al. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496. Link: https://goo.gl/8ssUrQ
- Winston F, Allis CD (1999) The bromodomain: a chromatin-targeting module? Nat Struct Biol 6: 601-604. Link: https://goo.gl/UaO4KK
- Zheng L, Zhou MM (2002) Bromodomain; an acetyl-lysine binding domain. FEBS Lett 513: 124–128. Link: https://goo.gl/lj0mr6
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, et al . (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120– 124. Link: https://goo.gl/YE4VP9
- Zhang Y, Reinberg D (2001) transcription Regulation by Histone Methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343-2360. Link: https://goo.gl/Wvu43t
- Shi Y (2007) Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 8: 829-833. Link: https://goo.gl/FJAHZT
- Becker PB, Horz W (2002) ATP dependent nucleosome remodeling. Annu Rev Biochem 71: 247-273. Link: https://goo.gl/CZygB8
- Kadam S, Emerson BM (2002) Mechanism of chromatin assembly and transcription. Curr Opin Cell Biol 14: 262-268. Link: https://goo.gl/JhwY9g
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887-905. Link: https://goo.gl/Djdtuu
- Thomä NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, et al. (2005) Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol 12: 350-356. Link: https://goo.gl/RV3Z0N
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475-487. Link: https://goo.gl/4ISclK
- Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD (1994) A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast, Proc Natl Acad Sci U S A 91: 1950–1954. Link: https://goo.gl/8xCnFK
- Peterson CL, Dingwal A, Scott MP (1994) Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional activation. Proc Natl Acad Sci USA 91: 2905-2908. Link: https://goo.gl/YpY9EU
- Côté J, Quinn J, Workman JL, Peterson CL (1994) Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53-60. Link: https://goo.gl/ACHg0m
- Sudarshanam P, Winston F (2000) The Swi/Snf family nucleosomeremodeling complexes and transcriptional control. Trends Genet 16: 345– 351. Link: https://goo.gl/3kKHiX
- Sawa H, Kouike H, Okano H (2000) Component of Swi/Snf complex are required for asymmetric cell division in C. elegans. Mol Cell 6: 617-624. Link: https://goo.gl/vkaLBE
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL (1996) DNA binding properties of the Yeast SWI/SNF complex. Nature 379: 844-847. Link: https://goo.gl/Su2BIR
- Utley RT, Côté J, Owen-Hughes T, Workman JL (1997) SWI/SNF Stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem 272: 12642-12649 Link: https://goo.gl/SNnAet
- Owen-Hughes T, Workman JL (1996) Remodelling of the hromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J 15: 4702-4712. Link: https://goo.gl/NzDKTZ
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, et al. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400: 784-787. Link: https://goo.gl/J5TaQL
- Raut VV, Pandey SM, Sainis JK (2011) Histone octamer trans-transfer: a signature mechanism of ATP-dependent chromatin remodelling unravelled in wheat nuclear extract. Annals of Botany 108: 1235–1246. Link: https://goo.gl/qGltS5
- de la Serna IL, Ohkawa Y, Higashi C, Dutta C, Osias J, et al. (2006) The microphthalmia transcription factor (Mitf) requires SWI/SNF enzymes to activate melanocyte specific genes. J Biol Chem 281: 20233-20241. Link: https://goo.gl/mFAf8T
- Han JA, Kim JI, Ongusaha PP, Hwang DH, Ballou LR, et al. (2002) P53-mediated induction of Cox-2 counteracts p53- or genotoxic stress-induced apoptosis. EMBO J 21: 5635-5644. Link: https://goo.gl/LVIDGr
- Patenge N, Elkin SK, Oettinger MA (2004) ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J Biol Chem 279: 35360-35367. Link: https://goo.gl/RmwwG1
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, et al. (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95: 625– 636. Link: https://goo.gl/lOki1O
- Rando OJ, Zhao K, Janmey P, Crabtree GR (2002) Phosphatidylinositol- dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci U S A 99: 2824– 2829. Link: https://goo.gl/h6Ufq9
- Hendricks KB, Shanahan F, Lees E (2004) Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol 24: 362-376. Link: https://goo.gl/mwgSdy
- Tsukiyama T, Wu C (1995) Purification and properties of an ATPdependent nucleosome remodeling factor. Cell 83: 1011–1020. Link: https://goo.gl/oUlrLH
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, et al. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598–602. Link:
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C (1999) Characterization of the Imitation Switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev 13: 686–697. Link: https://goo.gl/l5ToxV
- Aalfs JD, Narlikar GJ, Kingston RE (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J Biol Chem 276: 34270–34278. Link: https://goo.gl/wOLXa7
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, et al. (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20: 4892–4900. Link: https://goo.gl/zRrIQx
- Lazzaro MA, Picketts DJ (2001) Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem 77: 1145–1156. Link: https://goo.gl/NGsLP0
- Guschin D, Geiman TM, Kikyo N, Tremethick DJ, Wolffe AP, et al. (2000) Multiple ISWI ATPase complexes from Xenopus leavis: functional conservation of an ACF/CHRAC homolog. J Biol Chem 275: 35248-35255. Link: https://goo.gl/JMrUFL
- Grüne T, Brzeski J, Eberharter A, Clapier CR, Corona DF, et al. (2003) Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell: 12: 449–460. Link: https://goo.gl/F4JGfn
- Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23: 2715–2723. Link: https://goo.gl/PAbLd2
- Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, et al. (2002) Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10: 935–942. Link: https://goo.gl/DzA94Z
- Boyer LA, Latek RR, Peterson CL (2004) The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol 5: 158–163. Link: https://goo.gl/8ygyTB
- Brehm A, Längst G, Kehle J, Clapier CR, Imhof A, et al. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J 19: 4332– 4341. Link: https://goo.gl/xfBZj7
- Wang G, Ma A, Chow CM, Horsley D, Brown NR, et al. (2000) Conservation of Heterochromatin Protein 1 Function. Mol. Cell. Biol. 20: 6970– 6983. Link: https://goo.gl/vkWxWQ
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodeling complex. Nature 395: 917–921. Link: https://goo.gl/M7U1iC
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, et al. (1998) NURD, a Novel Complex with Both ATP-Dependent Chromatin-Remodeling and Histone Deacetylase Activities. Mol. Cell 2: 851–861. Link: https://goo.gl/gLSWtp
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The Dermatomyositis-Specific Autoantigen Mi2 Is a Component of a Complex Containing Histone Deacetylase and Nucleosome Remodeling Activities. Cell 95: 279– 289. Link: https://goo.gl/gQKzJA
- Wang HB, Zhang Y (2001) Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res 29: 2517– 2521. Link: https://goo.gl/wjpsQc
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, et al. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 23: 62– 66. Link: https://goo.gl/gDgBX4
- Wade PA, Jones PL, Vermaak D, Wolffe AP (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol 8: 843– 846. Link: https://goo.gl/KUWVDD
- Ebbert R, Birkmann A, Schüller HJ (1999) The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genesis part of a high-molecular-weight protein complex. Mol Microbiol 32: 741–751. Link: https://goo.gl/cFtR4z
- Shen X, Mizuguchi G, Hamiche A, Wu C (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature: 406: 541–544. Link: https://goo.gl/MSzReq
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, et al. (2005) A mammalian chromatin remodeling complex with similarities to the yeastINO80 complex. J Biol Chem 280: 41207–41212. Link: https://goo.gl/9Sl4QM
- Shen X, Ranallo R, Choi E, Wu C (2003) Involvement of actinrelated proteins in ATP-dependent chromatin remodeling. Mol Cell 12: 147–155. Link: https://goo.gl/CH1LMY
- van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. Link: https://goo.gl/G2WCg7
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA (2005) Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383. Link: https://goo.gl/zh68CV
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
